File conversion
lun. 22 octobre 2018 by Martin DeudonSummary
The Matlab code is on the eegFileConversion repository.
Conversion scripts are listed in the table below with their main function :
| Function | file | misc. | Down-sampling |
|---|---|---|---|
Format Conversion |
fileconv_nsx2edf() | Yes | |
| nsx2eeglab() | Yes | ||
| dirconv_nsx2edf | Yes | ||
Mono-to-bipolar |
fileconv_mono2bipolar_macro() | No | |
| fileconv_mono2bipolar_micro() | No | ||
| mono2bipolar_macro() | No | ||
| mono2bipolar_micro() | No | ||
| dirconv_mono2bipolar_macro | No | ||
| dirconv_mono2bipolar_micro | No | ||
Synchronization |
filesync_macromicro() | No | |
Divide |
filedivide_edf() | No | |
Downsampling |
fileconv_downsample_edf() | Yes | |
| dirconv_downsample_edf | Yes | ||
EpiFaR |
jediconv | Yes | |
| jediconv_dir | Yes | ||
| yodaconv | Experimental | Yes |
![]() All these scripts detect EEG channels based on the assumption that these channels'
name start with 'EEG '. This convention must be respected for the scripts to work
All these scripts detect EEG channels based on the assumption that these channels'
name start with 'EEG '. This convention must be respected for the scripts to work ![]()
In most of the scripts, you can specify the names of the bad channels, with the variable/argument badChannelNames which
must be a cell containing the names of the bad channels. These channels will be removed.
Format conversion
Blackrock NSx files
Blackrock files are saved in NS5/NEV format. The data are contained in the NS5 file, the NEV file can contain other information such as the triggers.
Matlab
To open the Blackrock file in Matlab, the NPMK toolbox is needed. It contains a lot a useful functions including openNSx() which allows to load NSx files into Matlab.
Open a NS5 file in Matlab with the fileconv_nsx2edf function. It writes an EDF file in the edf_dirpath and downsample the data by 10.
nsx_dir = 'C:\TestFiles\NSX';
nsx_filename = '20200410-144200-001.ns5';
edf_dirpath = 'C:\TestFiles\EDF';
downsampling_factor = 10;
[out_dirpath, out_filename, EEGs] = fileconv_nsx2edf(nsx_dir, nsx_filename, -1, downsampling_factor);
You can find a visual document showing the correspondencies between EEGLAB and Blackrock fields here
Python
In python you can use the neo package.
In python, there is no easy way to write data into the EDF file format (the pyedflib package is complex). The .fif format is a good alternative as it can be opened with micMac and with MNE.
Import a Blackrock file in Python with MNE and Neo :
import mne
import neo
import numpy as np
nsx_filepath = r'C:\TestFiles\NSX
bl = neo_loader.read(cascade=True, lazy=False)[0]
seg = bl.segments[0]
n_pnts, n_chan = len(seg.analogsignals[0]), len(seg.analogsignals)
ch_names = list()
data = np.zeros((n_chan, n_pnts), dtype=float)
for i, asig in enumerate(seg.analogsignals):
ch_names.append(asig.name)
# We need the ravel() here because Neo < 0.5 gave 1D, Neo 0.5 gives
# 2D (but still a single channel).
data[i, :] = asig.magnitude.ravel()
sfreq = int(seg.analogsignals[0].sampling_rate.magnitude)
info = mne.create_info(ch_names=ch_names, sfreq=sfreq)
raw = mne.io.RawArray(data, info)
Then you can save the raw MNE structure easily (in .fif format for example)
Neuralynx files
With Blackrock files (nsX), all the channels are stored in the same file and the file can be divided into several time periods. In Neuralynx format (ncs), each channel is stored in a separate file and can also be segmented into different time periods.
The eegFileConversion repository contain some scripts for converting Neuralynx files to EDF. You will need the NeuraLynx MATLAB Import/Export MEX Files
![]() These functions are a bit experimental and you should not rely too much on the ouput result
These functions are a bit experimental and you should not rely too much on the ouput result ![]()
With Neuralynx format, each channel is separated in a different file, and there may be several files for a single channel. The scripts differs in the following way :
CSCdir_to_EDF_divide: merge the channels together but do NOT merge the different files togetherCSCdir_to_EDF_merge: merge the channels together and merge also the different files together.CSCconv_writedatatoedf: function used by the two other scripts to contruct an EEGLAB structure from the data and convert it into an .edf file.
These scripts also separate the micro and the Macro channels which are in the same .ncs files. The way it discriminate micro and Macro channels is based on the channel name. It may not work if different naming convention are used !
New montage
The scripts and functions for creating a new montage use both EEGLAB and ERPLAB.
Macro files : monopolar to bipolar
The naming convention for bipolar files is to append '_b' to the file name
mono2bipolar_macro: main mono-to-bipolar conversion function. Take as argument and return an EEGLAB structure.fileconv_mono2bipolar_macro: wrapper around mono2bipolar_macro function. Write the bipolar .edf file.dirconv_mono2bipolar_macro: wrapper around mono2bipolar_macro function. Convert all .edf files in a directory into bipolar .edf files
Micro files
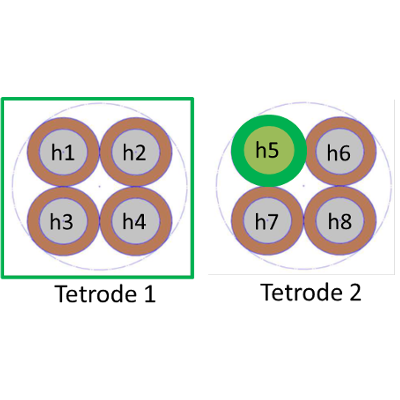
There are more possible montages for micro tetrodes that for macro electrodes. We have used mainly three montages so far :
| Reference commune (monopolar) | Inter-tetrode | Intra-tetrode |
|---|---|---|
 |
 |
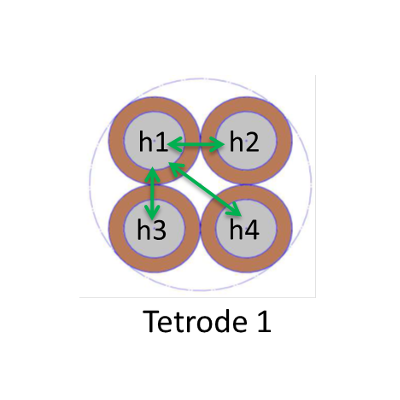 |
| Ref. is a macro-contact in the white matter | Ref. is a contact from another tetrode on the same electrode | Ref. is a contact from the same tetrode |
mono2bipolar_micro: main mono-to-bipolar conversion function. Takes as argument and returns an EEGLAB structure.fileconv_mono2bipolar_micro: wrapper around mono2bipolar_macro function. Write the bipolar .edf file.dirconv_mono2bipolar_micro: wrapper around mono2bipolar_macro function. Convert all .edf files in a directory into bipolar .edf files
The inter and intra tetrode montage has to be done manually. Modify the mono2bipolar_micro file, you have to specify the
tetrode names microNames, the number of channels per tetrode nChanPerMicro (8 or 12) and the bipolar montage bipolarMontage.
The bipolarMontage variable is a cell of vectors. Each vector defines the new channels for a single electrode.
case 34 % P34
microNames = {'b','tb','b'''};
nChanPerMicro = [12,8,12];
bipolarMontage = {[ 1,5;2,6;3,7;4,8;5,9;6,10;7,11;8,12;1,9;2,10;3,11;4,12],...
[ 1,5;2,6;3,7;4,8],...
[ 1,5;2,6;3,7;4,8;5,9;6,10;7,11;8,12;1,9;2,10;3,11;4,12]};
EpiFaR files
Several scripts are dedicated to the conversion of EpiFaR files and offer an automated pipeline :
- Convert the micro .ns5 file into an .edf, downsample it if needed and divide it into 10-minutes segments.
- Synchronize the Macro .edf file with the micro file. If the Macro file starts before, cut the beginning the macro file. If the Macro starts after, add a blank signal at the beginning of the macro file
- Divide the Macro synchronized file in 10-minutes segments
- Convert the Macro monopolar file into bipolar files
- Convert the micro monopolar file into bipolar files.
Some of these step are not mandatory and can be commented in the jediconv file. All the files starting with jediconv
deal with EpiFaR files. The yodaconv file can be used when the micro file is discontinued and composed of several parts.
However this script has not been tested thoroughly.